
 |
Rahul Kulkarni |
| Home | CV | Research | Publications | Group | Teaching |
|
A key focus of current research in the group is the interplay between stochastic gene expression and regulation by small RNAs. We combine different analytical tools (e.g. queueing theory and variational approaches) with stochastic simulations to model gene expression and its regulation. The models analyzed are, in part, based on discoveries made by combining our bioinformatic approaches with experimental validation by collaborators. We are interested in diverse applications of the formalism developed, ranging from modeling bacterial regulatory pathways (e.g. quorum-sensing and CsrA pathways) to proposing novel protocols for inferring molecular mechanisms of regulation from measurements of noise in gene expression. |
| Our research is currently funded by the following grants: |
| Previous research support: |
| Analytical framework for stochastic gene expression models |
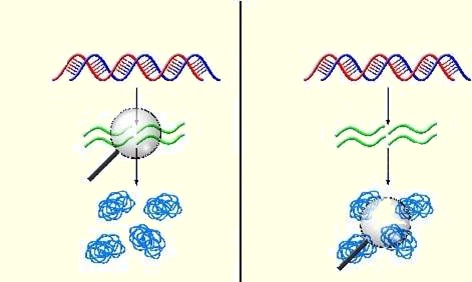 |
Recent research in our group has established a mapping between stochastic models of gene expression and systems analyzed in a branch of applied mathematics called queueing theory. By employing a famous theorem from queueing theory, Little's Law, we have derived exact results for general models of gene expression. The connection to queueing theory has also led us to derive new results relating to the noise in protein distributions that significantly extend current knowledge in the field. |
| Analytical framework for post-transcriptional regulation of gene expressions |
Our research has also focused on the interplay between regulation by sRNAs and stochastic gene expression. In previous work, a reaction scheme was introduced for regulation of gene expression by sRNAs via coupled stoichiometric degradation. While the exact solution of the corresponding stochastic model is analytically intractable, we were able to prove some exact results relating the means of the mRNA/sRNA probability distributions. For this model, in combination with the queueing-theory based analysis, we also showed how sRNAs can be used to quantify the degree of transcriptional bursting. Furthermore, we have recently developed a more general model of regulation by sRNAs. In the limit that fluctuations in sRNA concentration can be neglected, we obtained the exact analytical solution for this model. The results derived provide novel insights into the effects of different modes of post-transcriptional regulation on the noise in gene expression. Our future research will build on these studies to establish a general framework for regulation of gene expression and its applications. |
|
| Back To Top |
| Bioinformatic analysis of quorum-sensing and CsrA pathways |
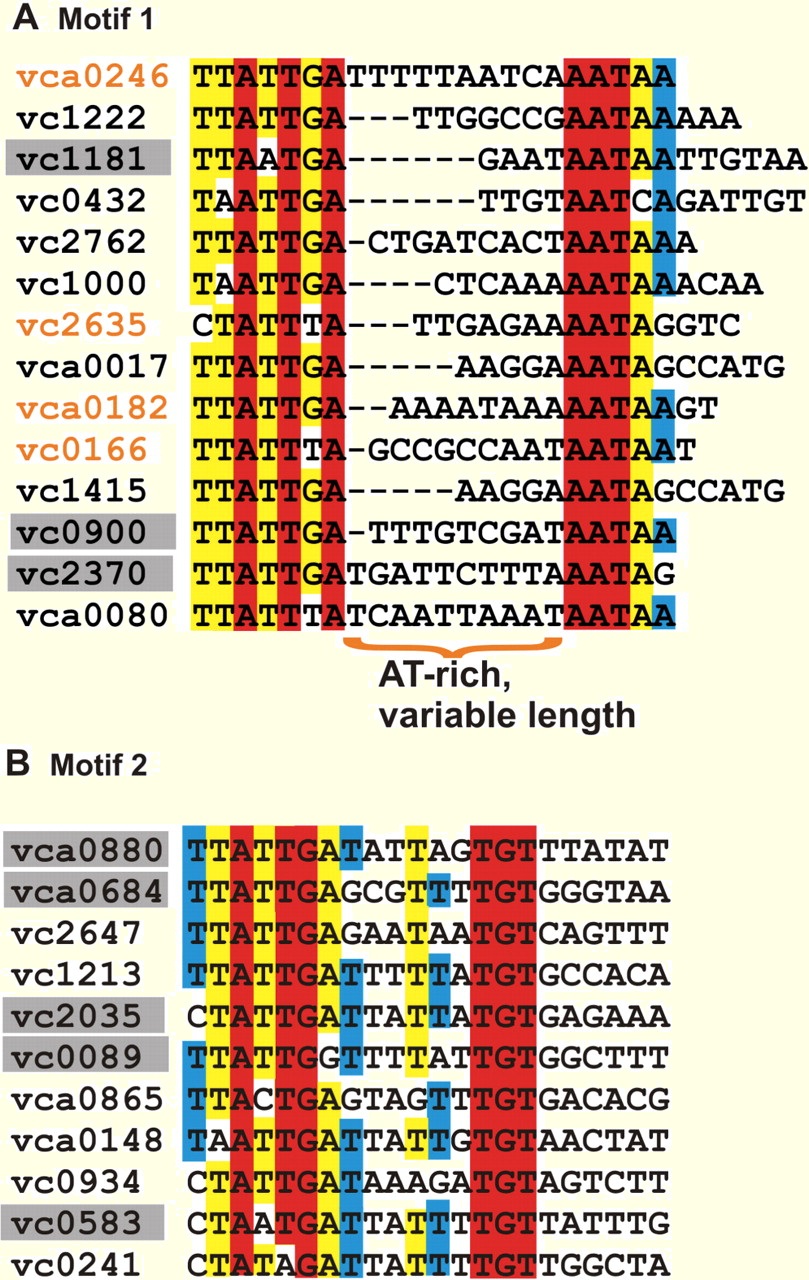 |
My collaboration with the groups of Bonnie Bassler (Princeton University) and Ned Wingreen (Princeton University) led to the discovery of small RNAs (sRNAs) in the QS pathways in Vibrio harveyi and Vibrio cholerae. In followup work with the Bassler group, we established links between the QS and CsrA pathways and discovered CsrA-regulating sRNAs in V. cholerae. Subsequently, my group developed a general bioinformatic code (CSRNA_FIND) for predicting CsrA-regulating sRNAs in diverse bacterial species. Our approach was able to identify all the previously discovered CsrA-regulating sRNAs and also make several novel predictions. For example, an open question in the field was identification of CsrA-regulating sRNAs in Legionella pneumophila. Our theoretical predictions for the corresponding sRNAs were recently validated by three different groups, thereby solving an outstanding problem. We have also developed bioinformatic approaches for significantly expanding current knowledge of regulons for bacterial transcription factors.
A case in point is the QS regulon in V. cholerae for which, in collaboration with the experimental group of Jun Zhu (U Penn), we have discovered several novel targets directly controlled by the QS master regulator HapR. |
| Back To Top |
| Applications to inverse problems in stochastic gene expression: |
Recent research focusing on gene network dynamics in single cells has shed new light on the processes underlying gene regulation. The increasing use of single-cell approaches underscores the need for developing theoretical tools for inference of underlying model parameters from experimental data. We plan to extend the analytical formalism developed for gene expression to formulate protocols for interfacing with single-cell data and estimating the underlying model parameters. |
| Applications to diverse cellular processes involving regulation of gene expression: |
Recent research on cellular processes, in particular involving microRNAs and sRNAs, has discovered many intriguing regulatory mechanisms controlling gene expression. While novel regulatory roles are increasingly being discovered, the corresponding modeling efforts which can provide quantitative insight have been lacking. We plan to address this gap in the field by a) identifying particularly important cellular processes involving regulation of noise in gene expression and b) developing and analyzing the corresponding stochastic models using the formalism we are developing. The initial analysis of these models will be integrated with teaching efforts. |
| Modeling of quorum-sensing and CsrA pathways: |
We plan to continue ongoing efforts to characterize and model global regulation by the QS and CsrA pathways. This work will be done in close collaboration with experimental colleagues. In collaboration with Steve Hagen (University of Florida) and Eric Stabb (University of Georgia), we plan to quantitatively model quorum-sensing in V. fischeri by integrating microfluidics-based single-cell measurements (Hagen), deterministic and stochastic modeling of the pathway (Kulkarni), and experimental microbiology (Stabb). Additionally, in collaboration with Jun Zhu (U Penn), we have made new discoveries relating to virulence regulation by ToxT in V. cholerae. These findings have opened the doors for further efforts at mapping out pathways and mechanisms responsible for V. cholerae virulence, which we plan to pursue. Furthermore, we have developed a computer code (CSRA_TARGET) for prediction of CsrA-regulated targets across bacterial genomes. We plan to further validate the predictions from our code in collaboration with the experimental group of Miguel Cámara (University of Nottingham), which will be a major step towards refinement and further development of the code. It is anticipated that the computational code developed will benefit research in multiple labs by providing testable predictions for CsrA-regulated target genes in diverse bacterial species. |
| Back To Top |